Structure-Based Drug-Design
SBDD 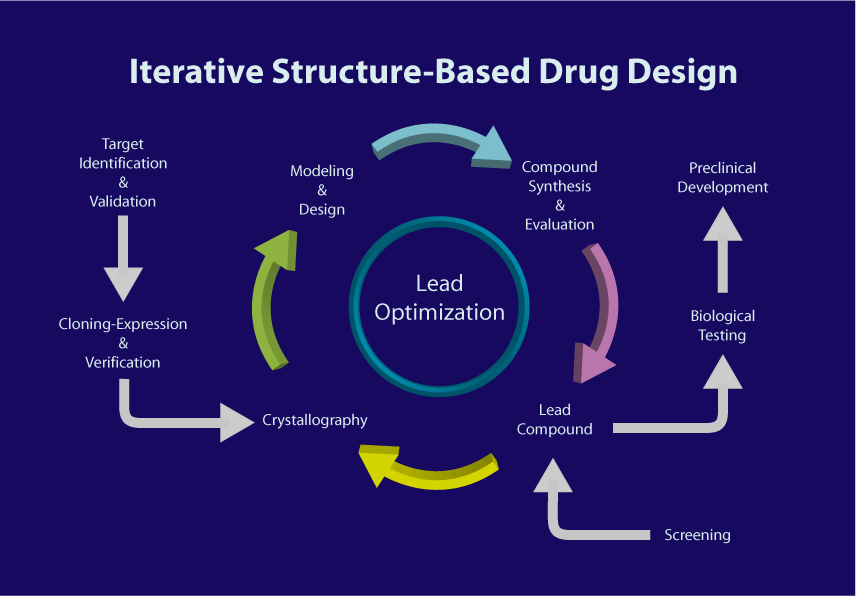 is an iterative process (Fig. 1), in which macromolecular crystallography has been the predominate technique used to elucidate the three-dimensional structure of drug targets. Although both nucleic acids and proteins are potential drug targets, by far the majority of such targets are proteins. Given that many proteins undergo considerable conformational change upon ligand binding, it is important to design drugs based on the crystallographic structures of protein-ligand complexes, not the unliganded structure.
is an iterative process (Fig. 1), in which macromolecular crystallography has been the predominate technique used to elucidate the three-dimensional structure of drug targets. Although both nucleic acids and proteins are potential drug targets, by far the majority of such targets are proteins. Given that many proteins undergo considerable conformational change upon ligand binding, it is important to design drugs based on the crystallographic structures of protein-ligand complexes, not the unliganded structure.
Crystallography has been successfully used in the de novo design of drugs, but its most important use has been, and will continue to be, in lead optimization (Figure below). It is important to note that what is being optimized is the affinity and specificity of compounds to their drug target. Lead optimization is a multi-step process that can be summarized as follow:
The process starts with cloning, expression and purification of the protein of interest. The protein is then crystallized in the presence of a ligand, which can be a non-hydrolysable substrate or can come from a biochemical or a cell-based screen. Ligands can also be low affinity compound fragments or scaffolds. The latter are generally a collection of basic chemical building blocks, each with a molecular weight of less than 200 Daltons. It is important to note that if the screen identifies several promising ligands, each with a unique scaffold, one should try to determine the structures of the drug target with as many of these as possible.
Once one or more liganded structures have been determined and refined, analysis of each structure will reveal sites on the ligand that can be optimized to enhance potency to the drug target. This can be accomplished by redesigning the ligand with greater hydrophobic, hydrogen-bonding and electrostatic complementarity to the molecular target. The design process can be simple and intuitive if one starts with a relatively high affinity lead. In this case only minor modifications to the existing ligand are introduced. Many of these modifications can be proposed from previous personal knowledge, or can be derived by computer modeling. There are numerous commercial and academic computer programs to aid in the analysis and design of new ligands. However, it is important to note that computational methods are still not reliable in predicting binding modes and affinities of ligands, mainly due to inaccuracies in force fields, limitations in dealing with ligand and target flexibility, the lack of reliable scoring functions, as well as the difficulties in treating solvent molecules. Therefore, even for seemingly minor modifications of the leads, it is still necessary to confirm the binding mode experimentally; there are countless examples in which the mode of binding significantly changes upon introduction of minor modifications to the original ligand.
Now that ligands have been designed, they should be chemically synthesized. It is prudent, if synthetically feasible and relatively easy, to synthesize a small library of five to ten compounds around the proposed ligand to obtain structure-activity relationship (SAR) data. Once the synthesized compounds are purified, they are tested in a relevant biochemical or cell-based assay to determine whether or not the design was successful. Occasionally a redesigned ligand will show less potency than the parent compound. Further cycles of structure determinations should reveal the reason.
The above three steps constitute one design cycle. It is often necessary to go through several iterations of the above cycle of structure determination, design, synthesis and testing before a drug candidate emerges (Fig. 1).
